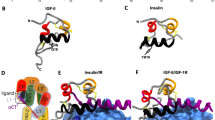
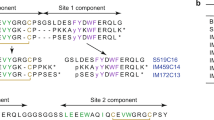
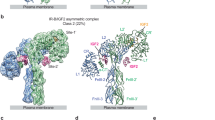
Abstract
Receptors of insulin and insulin-like growth factors (IGFs) are receptor tyrosine kinases whose signalling controls multiple aspects of animal physiology throughout life. In addition to regulating metabolism and growth, insulin–IGF receptor signalling has recently been linked to a variety of new, cell type-specific functions. In the last century, key questions have focused on how structural differences of insulin and IGFs affect receptor activation, and how insulin–IGF receptor signalling translates into pleiotropic biological functions. Technological advances such as cryo-electron microscopy have provided a detailed understanding of how native and engineered ligands activate insulin–IGF receptors. In this Review, we highlight recent structural and functional insights into the activation of insulin–IGF receptors, and summarize new agonists and antagonists developed for intervening in the activation of insulin–IGF receptor signalling. Furthermore, we discuss recently identified regulatory mechanisms beyond ligand–receptor interactions and functions of insulin–IGF receptor signalling in diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Petersen, M. C. & Shulman, G. I. Mechanisms of insulin action and insulin resistance. Physiol. Rev. 98, 2133–2223 (2018).
Santoro, A., McGraw, T. E. & Kahn, B. B. Insulin action in adipocytes, adipose remodeling, and systemic effects. Cell Metab. 33, 748–757 (2021).
Mathieu, C., Martens, P. J. & Vangoitsenhoven, R. One hundred years of insulin therapy. Nat. Rev. Endocrinol. 17, 715–725 (2021).
Boucher, J., Kleinridders, A. & Kahn, C. R. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb. Perspect. Biol. 6, a009191 (2014).
White, M. F. & Kahn, C. R. Insulin action at a molecular level — 100 years of progress. Mol. Metab. 52, 101304 (2021).
Kasuga, M., Karlsson, F. A. & Kahn, C. R. Insulin stimulates the phosphorylation of the 95,000-dalton subunit of its own receptor. Science 215, 185–187 (1982).
Haeusler, R. A., McGraw, T. E. & Accili, D. Biochemical and cellular properties of insulin receptor signalling. Nat. Rev. Mol. Cell Biol. 19, 31–44 (2018).
Kasuga, M., Zick, Y., Blithe, D. L., Crettaz, M. & Kahn, C. R. Insulin stimulates tyrosine phosphorylation of the insulin receptor in a cell-free system. Nature 298, 667–669 (1982).
White, M. F., Maron, R. & Kahn, C. R. Insulin rapidly stimulates tyrosine phosphorylation of a Mr-185,000 protein in intact cells. Nature 318, 183–186 (1985).
Sun, X. J. et al. Structure of the insulin receptor substrate IRS-1 defines a unique signal transduction protein. Nature 352, 73–77 (1991).
Siddle, K. Signalling by insulin and IGF receptors: supporting acts and new players. J. Mol. Endocrinol. 47, R1–R10 (2011).
De Meyts, P. in Endotext (eds Feingold, K. R. et al.) (MDText, 2016).
Accili, D. et al. Early neonatal death in mice homozygous for a null allele of the insulin receptor gene. Nat. Genet. 12, 106–109 (1996).
Joshi, R. L. et al. Targeted disruption of the insulin receptor gene in the mouse results in neonatal lethality. EMBO J. 15, 1542–1547 (1996).
Bruning, J. C. et al. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol. Cell 2, 559–569 (1998).
Lauro, D. et al. Impaired glucose tolerance in mice with a targeted impairment of insulin action in muscle and adipose tissue. Nat. Genet. 20, 294–298 (1998).
Bluher, M. et al. Adipose tissue selective insulin receptor knockout protects against obesity and obesity-related glucose intolerance. Dev. Cell 3, 25–38 (2002).
Michael, M. D. et al. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol. Cell 6, 87–97 (2000).
Kulkarni, R. N. et al. Tissue-specific knockout of the insulin receptor in pancreatic β cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell 96, 329–339 (1999).
Guerra, C. et al. Brown adipose tissue-specific insulin receptor knockout shows diabetic phenotype without insulin resistance. J. Clin. Invest. 108, 1205–1213 (2001).
Bruning, J. C. et al. Role of brain insulin receptor in control of body weight and reproduction. Science 289, 2122–2125 (2000).
Liu, J. P., Baker, J., Perkins, A. S., Robertson, E. J. & Efstratiadis, A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell 75, 59–72 (1993).
Kitamura, T. et al. Preserved pancreatic β-cell development and function in mice lacking the insulin receptor-related receptor. Mol. Cell Biol. 21, 5624–5630 (2001).
Nef, S. et al. Testis determination requires insulin receptor family function in mice. Nature 426, 291–295 (2003).
Barbieri, M., Bonafe, M., Franceschi, C. & Paolisso, G. Insulin/IGF-I-signaling pathway: an evolutionarily conserved mechanism of longevity from yeast to humans. Am. J. Physiol. Endocrinol. Metab. 285, E1064–E1071 (2003).
Lagueux, M., Lwoff, L., Meister, M., Goltzene, F. & Hoffmann, J. A. cDNAs from neurosecretory cells of brains of Locusta migratoria (Insecta, Orthoptera) encoding a novel member of the superfamily of insulins. Eur. J. Biochem. 187, 249–254 (1990).
Viola, C. M. et al. Structural conservation of insulin/IGF signalling axis at the insulin receptors level in Drosophila and humans. Nat. Commun. 14, 6271 (2023).
Smykal, V. et al. Complex evolution of insect insulin receptors and homologous decoy receptors, and functional significance of their multiplicity. Mol. Biol. Evol. 37, 1775–1789 (2020).
Altindis, E. et al. Viral insulin-like peptides activate human insulin and IGF-1 receptor signaling: a paradigm shift for host-microbe interactions. Proc. Natl Acad. Sci. USA 115, 2461–2466 (2018).
Ullrich, A. et al. Human insulin receptor and its relationship to the tyrosine kinase family of oncogenes. Nature 313, 756–761 (1985).
Ebina, Y. et al. The human insulin receptor cDNA: the structural basis for hormone-activated transmembrane signalling. Cell 40, 747–758 (1985).
Ullrich, A. et al. Insulin-like growth factor I receptor primary structure: comparison with insulin receptor suggests structural determinants that define functional specificity. EMBO J. 5, 2503–2512 (1986).
Shier, P. & Watt, V. M. Primary structure of a putative receptor for a ligand of the insulin family. J. Biol. Chem. 264, 14605–14608 (1989).
Lemmon, M. A. & Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 (2010).
Choi, E. & Bai, X. C. The activation mechanism of the insulin receptor: a structural perspective. Annu. Rev. Biochem. 92, 247–272 (2023).
Massague, J., Pilch, P. F. & Czech, M. P. Electrophoretic resolution of three major insulin receptor structures with unique subunit stoichiometries. Proc. Natl Acad. Sci. USA 77, 7137–7141 (1980).
Sparrow, L. G. et al. The disulfide bonds in the C-terminal domains of the human insulin receptor ectodomain. J. Biol. Chem. 272, 29460–29467 (1997).
Bravo, D. A., Gleason, J. B., Sanchez, R. I., Roth, R. A. & Fuller, R. S. Accurate and efficient cleavage of the human insulin proreceptor by the human proprotein-processing protease furin. Characterization and kinetic parameters using the purified, secreted soluble protease expressed by a recombinant baculovirus. J. Biol. Chem. 269, 25830–25837 (1994).
Bajaj, M., Waterfield, M. D., Schlessinger, J., Taylor, W. R. & Blundell, T. On the tertiary structure of the extracellular domains of the epidermal growth factor and insulin receptors. Biochim. Biophys. Acta 916, 220–226 (1987).
Schaffer, L. & Ljungqvist, L. Identification of a disulfide bridge connecting the α-subunits of the extracellular domain of the insulin receptor. Biochem. Biophys. Res. Commun. 189, 650–653 (1992).
Lawrence, M. C. Understanding insulin and its receptor from their three-dimensional structures. Mol. Metab. 52, 101255 (2021).
Li, J., Wu, J., Hall, C., Bai, X. C. & Choi, E. Molecular basis for the role of disulfide-linked αCTs in the activation of insulin-like growth factor 1 receptor and insulin receptor. eLife 11, e81286 (2022).
Rosenfeld, L. Insulin: discovery and controversy. Clin. Chem. 48, 2270–2288 (2002).
Banting, F. G., Best, C. H., Collip, J. B., Campbell, W. R. & Fletcher, A. A. Pancreatic extracts in the treatment of diabetes mellitus. 1922. Indian J. Med. Res. 125, 141–146 (2007).
Gorai, B. & Vashisth, H. Progress in simulation studies of insulin structure and function. Front. Endocrinol. 13, 908724 (2022).
Sanger, F. & Tuppy, H. The amino-acid sequence in the phenylalanyl chain of insulin. 1. The identification of lower peptides from partial hydrolysates. Biochem. J. 49, 463–481 (1951).
Sanger, F. & Tuppy, H. The amino-acid sequence in the phenylalanyl chain of insulin. 2. The investigation of peptides from enzymic hydrolysates. Biochem. J. 49, 481–490 (1951).
Adams, M. J. et al. Structure of rhombohedral 2 zinc insulin crystals. Nature 224, 491–495 (1969).
Mayer, J. P., Zhang, F. & DiMarchi, R. D. Insulin structure and function. Biopolymers 88, 687–713 (2007).
Weiss, M., Steiner, D. F. & Philipson, L. H. et al. in Endotext (eds Feingold, K. R. et al.) (MDText, 2014).
Palivec, V. et al. Computational and structural evidence for neurotransmitter-mediated modulation of the oligomeric states of human insulin in storage granules. J. Biol. Chem. 292, 8342–8355 (2017).
Mathieu, C., Gillard, P. & Benhalima, K. Insulin analogues in type 1 diabetes mellitus: getting better all the time. Nat. Rev. Endocrinol. 13, 385–399 (2017).
Omar-Hmeadi, M. & Idevall-Hagren, O. Insulin granule biogenesis and exocytosis. Cell Mol. Life Sci. 78, 1957–1970 (2021).
Salmon, W. D. Jr & Daughaday, W. H. A hormonally controlled serum factor which stimulates sulfate incorporation by cartilage in vitro. J. Lab. Clin. Med. 49, 825–836 (1957).
Daughaday, W. H. et al. Somatomedin: proposed designation for sulphation factor. Nature 235, 107 (1972).
Rinderknecht, E. & Humbel, R. E. Amino-terminal sequences of two polypeptides from human serum with nonsuppressible insulin-like and cell-growth-promoting activities: evidence for structural homology with insulin B chain. Proc. Natl Acad. Sci. USA 73, 4379–4381 (1976).
Terasawa, H. et al. Solution structure of human insulin-like growth factor II; recognition sites for receptors and binding proteins. EMBO J. 13, 5590–5597 (1994).
Nagao, H. et al. Distinct signaling by insulin and IGF-1 receptors and their extra- and intracellular domains. Proc. Natl Acad. Sci. USA 118, e2019474118 (2021).
Seino, S. & Bell, G. I. Alternative splicing of human insulin receptor messenger RNA. Biochem. Biophys. Res. Commun. 159, 312–316 (1989).
Frasca, F. et al. Insulin receptor isoform A, a newly recognized, high-affinity insulin-like growth factor II receptor in fetal and cancer cells. Mol. Cell Biol. 19, 3278–3288 (1999).
Belfiore, A. et al. Insulin receptor isoforms in physiology and disease: an updated view. Endocr. Rev. 38, 379–431 (2017).
Pandini, G. et al. Insulin/insulin-like growth factor I hybrid receptors have different biological characteristics depending on the insulin receptor isoform involved. J. Biol. Chem. 277, 39684–39695 (2002).
Slaaby, R. et al. Hybrid receptors formed by insulin receptor (IR) and insulin-like growth factor I receptor (IGF-IR) have low insulin and high IGF-1 affinity irrespective of the IR splice variant. J. Biol. Chem. 281, 25869–25874 (2006).
Benyoucef, S., Surinya, K. H., Hadaschik, D. & Siddle, K. Characterization of insulin/IGF hybrid receptors: contributions of the insulin receptor L2 and Fn1 domains and the alternatively spliced exon 11 sequence to ligand binding and receptor activation. Biochem. J. 403, 603–613 (2007).
Hexnerova, R. et al. Probing receptor specificity by sampling the conformational space of the insulin-like growth factor II C-domain. J. Biol. Chem. 291, 21234–21245 (2016).
Krizkova, K. et al. Insulin-insulin-like growth factors hybrids as molecular probes of hormone:receptor binding specificity. Biochemistry 55, 2903–2913 (2016).
Schaffer, L. A model for insulin binding to the insulin receptor. Eur. J. Biochem. 221, 1127–1132 (1994).
Bayne, M. L. et al. The C region of human insulin-like growth factor (IGF) I is required for high affinity binding to the type 1 IGF receptor. J. Biol. Chem. 264, 11004–11008 (1989).
Blyth, A. J., Kirk, N. S. & Forbes, B. E. Understanding IGF-II action through insights into receptor binding and activation. Cells 9, 2276 (2020).
Alvino, C. L. et al. A novel approach to identify two distinct receptor binding surfaces of insulin-like growth factor II. J. Biol. Chem. 284, 7656–7664 (2009).
Gauguin, L. et al. Alanine scanning of a putative receptor binding surface of insulin-like growth factor-I. J. Biol. Chem. 283, 20821–20829 (2008).
de Meyts, P., Roth, J., Neville, D. M. Jr, Gavin, J. R. III & Lesniak, M. A. Insulin interactions with its receptors: experimental evidence for negative cooperativity. Biochem. Biophys. Res. Commun. 55, 154–161 (1973).
Nielsen, J. et al. Structural investigations of full-length insulin receptor dynamics and signalling. J. Mol. Biol. 434, 167458 (2022).
Christoffersen, C. T. et al. Negative cooperativity in the insulin-like growth factor-I receptor and a chimeric IGF-I/insulin receptor. Endocrinology 135, 472–475 (1994).
De Meyts, P. & Whittaker, J. Structural biology of insulin and IGF1 receptors: implications for drug design. Nat. Rev. Drug Discov. 1, 769–783 (2002).
Croll, T. I. et al. Higher-resolution structure of the human insulin receptor ectodomain: multi-modal inclusion of the insert domain. Structure 24, 469–476 (2016).
Gutmann, T., Kim, K. H., Grzybek, M., Walz, T. & Coskun, U. Visualization of ligand-induced transmembrane signaling in the full-length human insulin receptor. J. Cell Biol. 217, 1643–1649 (2018).
Kohanski, R. A. Insulin receptor autophosphorylation. I. Autophosphorylation kinetics of the native receptor and its cytoplasmic kinase domain. Biochemistry 32, 5766–5772 (1993).
An, W. et al. Activation of the insulin receptor by insulin-like growth factor 2. Nat. Commun. 15, 2609 (2024).
Uchikawa, E., Choi, E., Shang, G., Yu, H. & Bai, X. C. Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex. eLife 8, e48630 (2019).
Gutmann, T. et al. Cryo-EM structure of the complete and ligand-saturated insulin receptor ectodomain. J. Cell Biol. 219, e201907210 (2020).
De Meyts, P. Insulin and its receptor: structure, function and evolution. Bioessays 26, 1351–1362 (2004).
Kristensen, C. et al. Alanine scanning mutagenesis of insulin. J. Biol. Chem. 272, 12978–12983 (1997).
Weis, F. et al. The signalling conformation of the insulin receptor ectodomain. Nat. Commun. 9, 4420 (2018).
Menting, J. G. et al. How insulin engages its primary binding site on the insulin receptor. Nature 493, 241–245 (2013).
Li, J. et al. Synergistic activation of the insulin receptor via two distinct sites. Nat. Struct. Mol. Biol. 29, 357–368 (2022).
Scapin, G. et al. Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis. Nature 556, 122–125 (2018).
Xiong, X. et al. Symmetric and asymmetric receptor conformation continuum induced by a new insulin. Nat. Chem. Biol. 18, 511–519 (2022).
Kertisova, A. et al. Insulin receptor Arg717 and IGF-1 receptor Arg704 play a key role in ligand binding and in receptor activation. Open Biol. 13, 230142 (2023).
Daly, M. E. et al. Acute effects on insulin sensitivity and diurnal metabolic profiles of a high-sucrose compared with a high-starch diet. Am. J. Clin. Nutr. 67, 1186–1196 (1998).
Shukla, A. P., Iliescu, R. G., Thomas, C. E. & Aronne, L. J. Food order has a significant impact on postprandial glucose and insulin levels. Diabetes Care 38, e98–e99 (2015).
Kanaley, J. A., Heden, T. D., Liu, Y. & Fairchild, T. J. Alteration of postprandial glucose and insulin concentrations with meal frequency and composition. Br. J. Nutr. 112, 1484–1493 (2014).
Wang, M., Li, J., Lim, G. E. & Johnson, J. D. Is dynamic autocrine insulin signaling possible? A mathematical model predicts picomolar concentrations of extracellular monomeric insulin within human pancreatic islets. PLoS ONE 8, e64860 (2013).
Najjar, S. M. & Perdomo, G. Hepatic insulin clearance: mechanism and physiology. Physiology 34, 198–215 (2019).
Xu, Y. et al. How ligand binds to the type 1 insulin-like growth factor receptor. Nat. Commun. 9, 821 (2018).
Zhang, X. et al. Cryo-EM studies of the apo states of human IGF1R. Biochem. Biophys. Res. Commun. 618, 148–152 (2022).
Li, J., Choi, E., Yu, H. & Bai, X. C. Structural basis of the activation of type 1 insulin-like growth factor receptor. Nat. Commun. 10, 4567 (2019).
Zhang, X. et al. Visualization of ligand-bound ectodomain assembly in the full-length human IGF-1 3eceptor by cryo-EM single-particle analysis. Structure 28, 555–561 (2020).
Surinya, K. H. et al. An investigation of the ligand binding properties and negative cooperativity of soluble insulin-like growth factor receptors. J. Biol. Chem. 283, 5355–5363 (2008).
Xu, Y. et al. How IGF-II binds to the human type 1 insulin-like growth factor receptor. Structure 28, 786–798 (2020).
Hakuno, F. & Takahashi, S. I. IGF1 receptor signaling pathways. J. Mol. Endocrinol. 61, T69–T86 (2018).
Xu, Y. et al. How insulin-like growth factor I binds to a hybrid insulin receptor type 1 insulin-like growth factor receptor. Structure 30, 1098–1108 (2022).
Kavran, J. M. et al. How IGF-1 activates its receptor. eLife 3, e03772 (2014).
Deyev, I. E. et al. Insulin receptor-related receptor as an extracellular alkali sensor. Cell Metab. 13, 679–689 (2011).
Petrenko, A. G., Zozulya, S. A., Deyev, I. E. & Eladari, D. Insulin receptor-related receptor as an extracellular pH sensor involved in the regulation of acid-base balance. Biochim. Biophys. Acta 1834, 2170–2175 (2013).
Clerk, A. & Sugden, P. H. The insulin receptor family in the heart: new light on old insights. Biosci. Rep. 42, BSR20221212 (2022).
Wang, L., Hall, C., Li, J., Choi, E. & Bai, X. C. Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor. Nat. Struct. Mol. Biol. 30, 661–669 (2023).
Suzawa, M. & Bland, M. L. Insulin signaling in development. Development 150, dev201599 (2023).
Duguay, S. J., Lai-Zhang, J. & Steiner, D. F. Mutational analysis of the insulin-like growth factor I prohormone processing site. J. Biol. Chem. 270, 17566–17574 (1995).
Komatsu, M., Takei, M., Ishii, H. & Sato, Y. Glucose-stimulated insulin secretion: a newer perspective. J. Diabetes Investig. 4, 511–516 (2013).
Henquin, J. C. Triggering and amplifying pathways of regulation of insulin secretion by glucose. Diabetes 49, 1751–1760 (2000).
Liu, J. L., Yakar, S. & LeRoith, D. Mice deficient in liver production of insulin-like growth factor I display sexual dimorphism in growth hormone-stimulated postnatal growth. Endocrinology 141, 4436–4441 (2000).
Vu, T. H. & Hoffman, A. R. Promoter-specific imprinting of the human insulin-like growth factor-II gene. Nature 371, 714–717 (1994).
Hou, J. C., Min, L. & Pessin, J. E. Insulin granule biogenesis, trafficking and exocytosis. Vitam. Horm. 80, 473–506 (2009).
Fang, Y. et al. Cytosolic pH is a direct nexus in linking environmental cues with insulin processing and secretion in pancreatic β cells. Cell Metab. 36, 1237–1251 (2024).
Argente, J., Chowen, J. A., Pérez-Jurado, L. A., Frystyk, J. & Oxvig, C. One level up: abnormal proteolytic regulation of IGF activity plays a role in human pathophysiology. EMBO Mol. Med. 9, 1338–1345 (2017).
Li, T. et al. TMED10 mediates the trafficking of insulin-like growth factor 2 along the secretory pathway for myoblast differentiation. Proc. Natl Acad. Sci. USA 120, e2215285120 (2023).
Allard, J. B. & Duan, C. IGF-binding proteins: why do they exist and why are there so many? Front. Endocrinol. 9, 117 (2018).
Baxter, R. C. & Martin, J. L. Structure of the Mr 140,000 growth hormone-dependent insulin-like growth factor binding protein complex: determination by reconstitution and affinity-labeling. Proc. Natl Acad. Sci. USA 86, 6898–6902 (1989).
Rajaram, S., Baylink, D. J. & Mohan, S. Insulin-like growth factor-binding proteins in serum and other biological fluids: regulation and functions. Endocr. Rev. 18, 801–831 (1997).
Baxter, R. C. Endocrine and cellular physiology and pathology of the insulin-like growth factor acid-labile subunit. Nat. Rev. Endocrinol. 20, 414–425 (2024).
Lewitt, M. S., Saunders, H., Phuyal, J. L. & Baxter, R. C. Complex formation by human insulin-like growth factor-binding protein-3 and human acid-labile subunit in growth hormone-deficient rats. Endocrinology 134, 2404–2409 (1994).
Guler, H. P., Zapf, J., Schmid, C. & Froesch, E. R. Insulin-like growth factors I and II in healthy man. Estimations of half-lives and production rates. Acta Endocrinol. 121, 753–758 (1989).
Yakar, S. et al. Serum complexes of insulin-like growth factor-1 modulate skeletal integrity and carbohydrate metabolism. FASEB J. 23, 709–719 (2009).
Guler, H. P., Zapf, J. & Froesch, E. R. Short-term metabolic effects of recombinant human insulin-like growth factor I in healthy adults. N. Engl. J. Med. 317, 137–140 (1987).
Dauber, A. et al. Mutations in pregnancy-associated plasma protein A2 cause short stature due to low IGF-I availability. EMBO Mol. Med. 8, 363–374 (2016).
Sitar, T., Popowicz, G. M., Siwanowicz, I., Huber, R. & Holak, T. A. Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins. Proc. Natl Acad. Sci. USA 103, 13028–13033 (2006).
Forbes, B. E. et al. Localization of an insulin-like growth factor (IGF) binding site of bovine IGF binding protein-2 using disulfide mapping and deletion mutation analysis of the C-terminal domain. J. Biol. Chem. 273, 4647–4652 (1998).
Neumann, G. M. & Bach, L. A. The N-terminal disulfide linkages of human insulin-like growth factor-binding protein-6 (hIGFBP-6) and hIGFBP-1 are different as determined by mass spectrometry. J. Biol. Chem. 274, 14587–14594 (1999).
Brinkman, A., Kortleve, D. J., Zwarthoff, E. C. & Drop, S. L. Mutations in the C-terminal part of insulin-like growth factor (IGF)-binding protein-1 result in dimer formation and loss of IGF binding capacity. Mol. Endocrinol. 5, 987–994 (1991).
Kim, H. et al. Structural basis for assembly and disassembly of the IGF/IGFBP/ALS ternary complex. Nat. Commun. 13, 4434 (2022).
Hsieh, T., Gordon, R. E., Clemmons, D. R., Busby, W. H. Jr. & Duan, C. Regulation of vascular smooth muscle cell responses to insulin-like growth factor (IGF)-I by local IGF-binding proteins. J. Biol. Chem. 278, 42886–42892 (2003).
Ren, H., Yin, P. & Duan, C. IGFBP-5 regulates muscle cell differentiation by binding to IGF-II and switching on the IGF-II auto-regulation loop. J. Cell Biol. 182, 979–991 (2008).
Zhang, C. et al. IGF binding protein-6 expression in vascular endothelial cells is induced by hypoxia and plays a negative role in tumor angiogenesis. Int. J. Cancer 130, 2003–2012 (2012).
Imai, Y. et al. Protease-resistant form of insulin-like growth factor-binding protein 5 is an inhibitor of insulin-like growth factor-I actions on porcine smooth muscle cells in culture. J. Clin. Invest. 100, 2596–2605 (1997).
Conover, C. A. et al. Metalloproteinase pregnancy-associated plasma protein A is a critical growth regulatory factor during fetal development. Development 131, 1187–1194 (2004).
Liu, C. et al. The metalloproteinase Papp-aa controls epithelial cell quiescence-proliferation transition. eLife 9, e52322 (2020).
Marouli, E. et al. Rare and low-frequency coding variants alter human adult height. Nature 542, 186–190 (2017).
Conover, C. A. & Oxvig, C. The pregnancy-associated plasma protein-A (PAPP-A) story. Endocr. Rev. 44, 1012–1028 (2023).
Lawrence, J. B. et al. The insulin-like growth factor (IGF)-dependent IGF binding protein-4 protease secreted by human fibroblasts is pregnancy-associated plasma protein-A. Proc. Natl Acad. Sci. USA 96, 3149–3153 (1999).
Boldt, H. B. et al. The Lin12–Notch repeats of pregnancy-associated plasma protein-A bind calcium and determine its proteolytic specificity. J. Biol. Chem. 279, 38525–38531 (2004).
Laursen, L. S. et al. Cell surface targeting of pregnancy-associated plasma protein A proteolytic activity. Reversible adhesion is mediated by two neighboring short consensus repeats. J. Biol. Chem. 277, 47225–47234 (2002).
Judge, R. A. et al. Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition. Nat. Commun. 13, 5500 (2022).
Kobberø, S. D. et al. Structure of the proteolytic enzyme PAPP-A with the endogenous inhibitor stanniocalcin-2 reveals its inhibitory mechanism. Nat. Commun. 13, 6084 (2022).
Sridar, J. et al. Cryo-EM structure of human PAPP-A2 and mechanism of substrate recognition. Commun. Chem. 6, 234 (2023).
Zhong, Q. et al. Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2. Cell Discov. 8, 137 (2022).
Oxvig, C. & Conover, C. A. The stanniocalcin–PAPP-A–IGFBP–IGF axis. J. Clin. Endocrinol. Metab. 108, 1624–1633 (2023).
Hall, C., Yu, H. & Choi, E. Insulin receptor endocytosis in the pathophysiology of insulin resistance. Exp. Mol. Med. 52, 911–920 (2020).
Wu, J., Park, S. H. & Choi, E. The insulin receptor endocytosis. Prog. Mol. Biol. Transl. Sci. 194, 79–107 (2023).
Chen, Y., Huang, L., Qi, X. & Chen, C. Insulin receptor trafficking: consequences for insulin sensitivity and diabetes. Int. J. Mol. Sci. 20, 5007 (2019).
Park, J. et al. MAD2-dependent insulin receptor endocytosis regulates metabolic homeostasis. Diabetes 72, 1781–1794 (2023).
Najjar, S. M., Caprio, S. & Gastaldelli, A. Insulin clearance in health and disease. Annu. Rev. Physiol. 85, 363–381 (2023).
Choi, E. et al. Mitotic regulators and the SHP2–MAPK pathway promote IR endocytosis and feedback regulation of insulin signaling. Nat. Commun. 10, 1473 (2019).
Choi, E., Zhang, X., Xing, C. & Yu, H. Mitotic checkpoint regulators control insulin signaling and metabolic homeostasis. Cell 166, 567–581 (2016).
Yoneyama, Y. et al. IRS-1 acts as an endocytic regulator of IGF-I receptor to facilitate sustained IGF signaling. eLife 7, e32893 (2018).
Poy, M. N. et al. CEACAM1 regulates insulin clearance in liver. Nat. Genet. 30, 270–276 (2002).
Soni, P., Lakkis, M., Poy, M. N., Fernstrom, M. A. & Najjar, S. M. The differential effects of pp120 (Ceacam 1) on the mitogenic action of insulin and insulin-like growth factor 1 are regulated by the nonconserved tyrosine 1316 in the insulin receptor. Mol. Cell Biol. 20, 3896–3905 (2000).
Grandl, G. et al. Global, neuronal or β cell-specific deletion of inceptor improves glucose homeostasis in male mice with diet-induced obesity. Nat. Metab. 6, 448–457 (2024).
Ansarullah et al. Inceptor counteracts insulin signalling in β-cells to control glycaemia. Nature 590, 326–331 (2021).
Liu, X. et al. Insulin induces insulin receptor degradation in the liver through EphB4. Nat. Metab. 4, 1202–1213 (2022).
Fagerholm, S., Ortegren, U., Karlsson, M., Ruishalme, I. & Stralfors, P. Rapid insulin-dependent endocytosis of the insulin receptor by caveolae in primary adipocytes. PLoS ONE 4, e5985 (2009).
Yamakawa, D. et al. Primary cilia-dependent lipid raft/caveolin dynamics regulate adipogenesis. Cell Rep. 34, 108817 (2021).
Gustavsson, J. et al. Localization of the insulin receptor in caveolae of adipocyte plasma membrane. FASEB J. 13, 1961–1971 (1999).
Kabayama, K. et al. Dissociation of the insulin receptor and caveolin-1 complex by ganglioside GM3 in the state of insulin resistance. Proc. Natl Acad. Sci. USA 104, 13678–13683 (2007).
Nystrom, F. H., Chen, H., Cong, L. N., Li, Y. & Quon, M. J. Caveolin-1 interacts with the insulin receptor and can differentially modulate insulin signaling in transfected Cos-7 cells and rat adipose cells. Mol. Endocrinol. 13, 2013–2024 (1999).
Imamura, T. et al. Two naturally occurring mutations in the kinase domain of insulin receptor accelerate degradation of the insulin receptor and impair the kinase activity. J. Biol. Chem. 269, 31019–31027 (1994).
Hancock, M. L. et al. Insulin receptor associates with promoters genome-wide and regulates gene expression. Cell 177, 722–736 (2019).
Aleksic, T. et al. Type 1 insulin-like growth factor receptor translocates to the nucleus of human tumor cells. Cancer Res. 70, 6412–6419 (2010).
Packham, S. et al. Nuclear translocation of IGF-1R via p150Glued and an importin-β/RanBP2-dependent pathway in cancer cells. Oncogene 34, 2227–2238 (2015).
Dall’Agnese, A. et al. The dynamic clustering of insulin receptor underlies its signaling and is disrupted in insulin resistance. Nat. Commun. 13, 7522 (2022).
Butkowski, E. G. & Jelinek, H. F. Hyperglycaemia, oxidative stress and inflammatory markers. Redox Rep. 22, 257–264 (2017).
Zhou, K. et al. Spatiotemporal regulation of insulin signaling by liquid-liquid phase separation. Cell Discov. 8, 64 (2022).
Gao, X. K. et al. Phase separation of insulin receptor substrate 1 drives the formation of insulin/IGF-1 signalosomes. Cell Discov. 8, 60 (2022).
Ahn, M. Y., Katsanakis, K. D., Bheda, F. & Pillay, T. S. Primary and essential role of the adaptor protein APS for recruitment of both c-Cbl and its associated protein CAP in insulin signaling. J. Biol. Chem. 279, 21526–21532 (2004).
Monami, G., Emiliozzi, V. & Morrione, A. Grb10/Nedd4-mediated multiubiquitination of the insulin-like growth factor receptor regulates receptor internalization. J. Cell Physiol. 216, 426–437 (2008).
Vecchione, A., Marchese, A., Henry, P., Rotin, D. & Morrione, A. The Grb10/Nedd4 complex regulates ligand-induced ubiquitination and stability of the insulin-like growth factor I receptor. Mol. Cell Biol. 23, 3363–3372 (2003).
Song, R. et al. Central role of E3 ubiquitin ligase MG53 in insulin resistance and metabolic disorders. Nature 494, 375–379 (2013).
Girnita, L. et al. β-arrestin and Mdm2 mediate IGF-1 receptor-stimulated ERK activation and cell cycle progression. J. Biol. Chem. 282, 11329–11338 (2007).
Girnita, L., Girnita, A. & Larsson, O. Mdm2-dependent ubiquitination and degradation of the insulin-like growth factor 1 receptor. Proc. Natl Acad. Sci. USA 100, 8247–8252 (2003).
Nagarajan, A. et al. MARCH1 regulates insulin sensitivity by controlling cell surface insulin receptor levels. Nat. Commun. 7, 12639 (2016).
Tawo, R. et al. The ubiquitin ligase CHIP integrates proteostasis and aging by regulation of insulin receptor turnover. Cell 169, 470–482 (2017).
Zhou, H. L. et al. An enzyme that selectively S-nitrosylates proteins to regulate insulin signaling. Cell 186, 5812–5825 (2023).
Sehat, B. et al. SUMOylation mediates the nuclear translocation and signaling of the IGF-1 receptor. Sci. Signal. 3, ra10 (2010).
van Meer, G., Voelker, D. R. & Feigenson, G. W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 (2008).
Pilon, M. Revisiting the membrane-centric view of diabetes. Lipids Health Dis. 15, 167 (2016).
Stubbs, C. D. & Smith, A. D. The modification of mammalian membrane polyunsaturated fatty acid composition in relation to membrane fluidity and function. Biochim. Biophys. Acta 779, 89–137 (1984).
Ferrara, P. J. et al. Lysophospholipid acylation modulates plasma membrane lipid organization and insulin sensitivity in skeletal muscle. J. Clin. Invest. 131, e135963 (2021).
Mitrofanova, A. et al. SMPDL3b modulates insulin receptor signaling in diabetic kidney disease. Nat. Commun. 10, 2692 (2019).
McElroy, B., Powell, J. C. & McCarthy, J. V. The insulin-like growth factor 1 (IGF-1) receptor is a substrate for γ-secretase-mediated intramembrane proteolysis. Biochem. Biophys. Res. Commun. 358, 1136–1141 (2007).
Kasuga, K., Kaneko, H., Nishizawa, M., Onodera, O. & Ikeuchi, T. Generation of intracellular domain of insulin receptor tyrosine kinase by γ-secretase. Biochem. Biophys. Res. Commun. 360, 90–96 (2007).
Meakin, P. J. et al. The β secretase BACE1 regulates the expression of insulin receptor in the liver. Nat. Commun. 9, 1306 (2018).
Guo, X. et al. Regulation of age-associated insulin resistance by MT1-MMP-mediated cleavage of insulin receptor. Nat. Commun. 13, 3749 (2022).
Kim, K. et al. γ-Secretase inhibition lowers plasma triglyceride-rich lipoproteins by stabilizing the LDL receptor. Cell Metab. 27, 816–827 (2018).
Fujita, M., Takada, Y. K. & Takada, Y. Insulin-like growth factor (IGF) signaling requires αvβ3–IGF1–IGF type 1 receptor (IGF1R) ternary complex formation in anchorage independence, and the complex formation does not require IGF1R and Src activation. J. Biol. Chem. 288, 3059–3069 (2013).
Beauvais, D. M. & Rapraeger, A. C. Syndecan-1 couples the insulin-like growth factor-1 receptor to inside-out integrin activation. J. Cell Sci. 123, 3796–3807 (2010).
Clemmons, D. R. & Maile, L. A. Interaction between insulin-like growth factor-I receptor and αVβ3 integrin linked signaling pathways: cellular responses to changes in multiple signaling inputs. Mol. Endocrinol. 19, 1–11 (2005).
Tahimic, C. G. et al. Regulation of ligand and shear stress-induced insulin-like growth factor 1 (IGF1) signaling by the integrin pathway. J. Biol. Chem. 291, 8140–8149 (2016).
Saegusa, J. et al. The direct binding of insulin-like growth factor-1 (IGF-1) to integrin αvβ3 is involved in IGF-1 signaling. J. Biol. Chem. 284, 24106–24114 (2009).
Oh, Y. et al. Synthesis and characterization of insulin-like growth factor-binding protein (IGFBP)-7. Recombinant human mac25 protein specifically binds IGF-I and -II. J. Biol. Chem. 271, 30322–30325 (1996).
Grotendorst, G. R., Lau, L. F. & Perbal, B. CCN proteins are distinct from and should not be considered members of the insulin-like growth factor-binding protein superfamily. Endocrinology 141, 2254–2256 (2000).
Evdokimova, V. et al. IGFBP7 binds to the IGF-1 receptor and blocks its activation by insulin-like growth factors. Sci. Signal. 5, ra92 (2012).
Zhang, L. et al. Insulin-like growth factor-binding protein-7 (IGFBP7) links senescence to heart failure. Nat. Cardiovasc. Res. 1, 1195–1214 (2022).
Artico, L. L. et al. Physiologic IGFBP7 levels prolong IGF1R activation in acute lymphoblastic leukemia. Blood Adv. 5, 3633–3646 (2021).
Davies, B. S., Fong, L. G., Yang, S. H., Coffinier, C. & Young, S. G. The posttranslational processing of prelamin A and disease. Annu. Rev. Genomics Hum. Genet. 10, 153–174 (2009).
Jiang, B. et al. Progerin modulates the IGF-1R/Akt signaling involved in aging. Sci. Adv. 8, eabo0322 (2022).
Liu, S. Y. & Ikegami, K. Nuclear lamin phosphorylation: an emerging role in gene regulation and pathogenesis of laminopathies. Nucleus 11, 299–314 (2020).
Worman, H. J., Fong, L. G., Muchir, A. & Young, S. G. Laminopathies and the long strange trip from basic cell biology to therapy. J. Clin. Invest. 119, 1825–1836 (2009).
Safavi-Hemami, H. et al. Specialized insulin is used for chemical warfare by fish-hunting cone snails. Proc. Natl Acad. Sci. USA 112, 1743–1748 (2015).
Ahorukomeye, P. et al. Fish-hunting cone snail venoms are a rich source of minimized ligands of the vertebrate insulin receptor. eLife 8, e41574 (2019).
Menting, J. G. et al. A minimized human insulin-receptor-binding motif revealed in a Conus geographus venom insulin. Nat. Struct. Mol. Biol. 23, 916–920 (2016).
Bao, S. J., Xie, D. L., Zhang, J. P., Chang, W. R. & Liang, D. C. Crystal structure of desheptapeptide(B24-B30)insulin at 1.6 Å resolution: implications for receptor binding. Proc. Natl Acad. Sci. USA 94, 2975–2980 (1997).
Xiong, X. et al. A structurally minimized yet fully active insulin based on cone-snail venom insulin principles. Nat. Struct. Mol. Biol. 27, 615–624 (2020).
Chrudinova, M. et al. Characterization of viral insulins reveals white adipose tissue-specific effects in mice. Mol. Metab. 44, 101121 (2021).
Zhang, F., Altindis, E., Kahn, C. R., DiMarchi, R. D. & Gelfanov, V. A viral insulin-like peptide is a natural competitive antagonist of the human IGF-1 receptor. Mol. Metab. 53, 101316 (2021).
Moreau, F. et al. Interaction of a viral insulin-like peptide with the IGF-1 receptor produces a natural antagonist. Nat. Commun. 13, 6700 (2022).
Belavgeni, A. et al. vPIF-1 is an insulin-like antiferroptotic viral peptide. Proc. Natl Acad. Sci. USA 120, e2300320120 (2023).
Wu, M. et al. Author correction: Functionally selective signaling and broad metabolic benefits by novel insulin receptor partial agonists. Nat. Commun. 15, 688 (2024).
Chen, Y. S. et al. Insertion of a synthetic switch into insulin provides metabolite-dependent regulation of hormone-receptor activation. Proc. Natl Acad. Sci. USA 118, e2103518118 (2021).
Liu, Y. et al. Recent progress in glucose-responsive insulin. Diabetes 73, 1377–1388 (2024).
Hoeg-Jensen, T. Review: glucose-sensitive insulin. Mol. Metab. 46, 101107 (2021).
Hoeg-Jensen, T. et al. Glucose-sensitive insulin with attenuation of hypoglycaemia. Nature 634, 944–951 (2024).
Brange, J., Andersen, L., Laursen, E. D., Meyn, G. & Rasmussen, E. Toward understanding insulin fibrillation. J. Pharm. Sci. 86, 517–525 (1997).
Hua, Q. X. & Weiss, M. A. Mechanism of insulin fibrillation: the structure of insulin under amyloidogenic conditions resembles a protein-folding intermediate. J. Biol. Chem. 279, 21449–21460 (2004).
Wang, L. et al. Structural basis of insulin fibrillation. Sci. Adv. 9, eadi1057 (2023).
Pillutla, R. C. et al. Peptides identify the critical hotspots involved in the biological activation of the insulin receptor. J. Biol. Chem. 277, 22590–22594 (2002).
Schaffer, L. et al. Assembly of high-affinity insulin receptor agonists and antagonists from peptide building blocks. Proc. Natl Acad. Sci. USA 100, 4435–4439 (2003).
Jensen, M., Hansen, B., De Meyts, P., Schaffer, L. & Urso, B. Activation of the insulin receptor by insulin and a synthetic peptide leads to divergent metabolic and mitogenic signaling and responses. J. Biol. Chem. 282, 35179–35186 (2007).
Park, J. et al. Activation of the insulin receptor by an insulin mimetic peptide. Nat. Commun. 13, 5594 (2022).
Kirk, N. S. et al. Activation of the human insulin receptor by non-insulin-related peptides. Nat. Commun. 13, 5695 (2022).
Lawrence, C. F. et al. Insulin mimetic peptide disrupts the primary binding site of the insulin receptor. J. Biol. Chem. 291, 15473–15481 (2016).
Keefe, A. D., Pai, S. & Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 9, 537–550 (2010).
Yunn, N. O. et al. An aptamer agonist of the insulin receptor acts as a positive or negative allosteric modulator, depending on its concentration. Exp. Mol. Med. 54, 531–541 (2022).
Yunn, N. O. et al. A hotspot for enhancing insulin receptor activation revealed by a conformation-specific allosteric aptamer. Nucleic Acids Res. 49, 700–712 (2021).
Kim, J. et al. Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures. Nat. Commun. 13, 6500 (2022).
Lee, J., Miyazaki, M., Romeo, G. R. & Shoelson, S. E. Insulin receptor activation with transmembrane domain ligands. J. Biol. Chem. 289, 19769–19777 (2014).
Bhaskar, V. et al. A fully human, allosteric monoclonal antibody that activates the insulin receptor and improves glycemic control. Diabetes 61, 1263–1271 (2012).
Spratt, J. et al. Multivalent insulin receptor activation using insulin-DNA origami nanostructures. Nat. Nanotechnol. 19, 237–245 (2024).
LeRoith, D., Holly, J. M. P. & Forbes, B. E. Insulin-like growth factors: ligands, binding proteins, and receptors. Mol. Metab. 52, 101245 (2021).
Damiola, F. et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 14, 2950–2961 (2000).
Stenvers, D. J., Scheer, F., Schrauwen, P., la Fleur, S. E. & Kalsbeek, A. Circadian clocks and insulin resistance. Nat. Rev. Endocrinol. 15, 75–89 (2019).
Crosby, P. et al. Insulin/IGF-1 drives PERIOD synthesis to entrain circadian rhythms with feeding time. Cell 177, 896–909 (2019).
González-Vila, A. et al. Astrocytic insulin receptor controls circadian behavior via dopamine signaling in a sexually dimorphic manner. Nat. Commun. 14, 8175 (2023).
Fougeray, T. et al. The hepatocyte insulin receptor is required to program the liver clock and rhythmic gene expression. Cell Rep. 39, 110674 (2022).
Ziegler, A. N. et al. Insulin-like growth factor II: an essential adult stem cell niche constituent in brain and intestine. Stem Cell Rep. 12, 816–830 (2019).
Becker, C., Lust, K. & Wittbrodt, J. Igf signaling couples retina growth with body growth by modulating progenitor cell division. Development 148, dev199133 (2021).
Kamei, H. & Duan, C. Alteration of organ size and allometric scaling by organ-specific targeting of IGF signaling. Gen. Comp. Endocrinol. 314, 113922 (2021).
Tu, X. et al. Local autocrine plasticity signaling in single dendritic spines by insulin-like growth factors. Sci. Adv. 9, eadg0666 (2023).
Pandey, K. et al. Neuronal activity drives IGF2 expression from pericytes to form long-term memory. Neuron 111, 3819–3836 (2023).
Salazar-Petres, E. R. & Sferruzzi-Perri, A. N. Pregnancy-induced changes in β-cell function: what are the key players? J. Physiol. 600, 1089–1117 (2022).
Lopez-Tello, J. et al. Fetal manipulation of maternal metabolism is a critical function of the imprinted Igf2 gene. Cell Metab. 35, 1195–1208 (2023).
Sandovici, I. et al. The imprinted Igf2–Igf2r axis is critical for matching placental microvasculature expansion to fetal growth. Dev. Cell 57, 63–79 (2022).
Yang, Y. et al. Endogenous IGF signaling directs heterogeneous mesoderm differentiation in human embryonic stem cells. Cell Rep. 29, 3374–3384 (2019).
Wamaitha, S. E. et al. IGF1-mediated human embryonic stem cell self-renewal recapitulates the embryonic niche. Nat. Commun. 11, 764 (2020).
Okawa, E. R. et al. Essential roles of insulin and IGF-1 receptors during embryonic lineage development. Mol. Metab. 47, 101164 (2021).
Chell, J. M. & Brand, A. H. Nutrition-responsive glia control exit of neural stem cells from quiescence. Cell 143, 1161–1173 (2010).
Sousa-Nunes, R., Yee, L. L. & Gould, A. P. Fat cells reactivate quiescent neuroblasts via TOR and glial insulin relays in Drosophila. Nature 471, 508–512 (2011).
Lozano-Ureña, A. et al. IGF2 interacts with the imprinted gene Cdkn1c to promote terminal differentiation of neural stem cells. Development 150, dev200563 (2023).
Venkatraman, A. et al. Maternal imprinting at the H19–Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature 500, 345–349 (2013).
Lu, J. et al. IGFBP1 increases β-cell regeneration by promoting α- to β-cell transdifferentiation. EMBO J. 35, 2026–2044 (2016).
Liu, C. et al. Ca2+ concentration-dependent premature death of igfbp5a−/− fish reveals a critical role of IGF signaling in adaptive epithelial growth. Sci. Signal. 11, eaat2231 (2018).
Li, Y. et al. ROS signaling-induced mitochondrial Sgk1 expression regulates epithelial cell renewal. Proc. Natl Acad. Sci. USA 120, e2216310120 (2023).
Wang, J. et al. Bone marrow-derived IGF-1 orchestrates maintenance and regeneration of the adult skeleton. Proc. Natl Acad. Sci. USA 120, e2203779120 (2023).
Xin, M. et al. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci. Signal. 4, ra70 (2011).
Holly, J. M. P., Biernacka, K. & Perks, C. M. The neglected insulin: IGF-II, a metabolic regulator with implications for diabetes, obesity, and cancer. Cells 8, 1207 (2019).
Garla, V. et al. Non-islet cell hypoglycemia: case series and review of the literature. Front. Endocrinol. 10, 316 (2019).
Fang, F., Goldstein, J. L., Shi, X., Liang, G. & Brown, M. S. Unexpected role for IGF-1 in starvation: maintenance of blood glucose. Proc. Natl Acad. Sci. USA 119, e2208855119 (2022).
Douglas, R. S. et al. Teprotumumab for the treatment of active thyroid eye disease. N. Engl. J. Med. 382, 341–352 (2020).
Freychet, P., Roth, J. & Neville, D. M. Jr. Insulin receptors in the liver: specific binding of [125 I]insulin to the plasma membrane and its relation to insulin bioactivity. Proc. Natl Acad. Sci. USA 68, 1833–1837 (1971).
White, M. F., Shoelson, S. E., Keutmann, H. & Kahn, C. R. A cascade of tyrosine autophosphorylation in the β-subunit activates the phosphotransferase of the insulin receptor. J. Biol. Chem. 263, 2969–2980 (1988).
Rajagopalan, M., Neidigh, J. L. & McClain, D. A. Amino acid sequences Gly-Pro-Leu-Tyr and Asn-Pro-Glu-Tyr in the submembranous domain of the insulin receptor are required for normal endocytosis. J. Biol. Chem. 266, 23068–23073 (1991).
Backer, J. M. et al. Phosphatidylinositol 3′-kinase is activated by association with IRS-1 during insulin stimulation. EMBO J. 11, 3469–3479 (1992).
Cantley, L. C. The phosphoinositide 3-kinase pathway. Science 296, 1655–1657 (2002).
Manning, B. D. & Toker, A. AKT/PKB signaling: navigating the network. Cell 169, 381–405 (2017).
Sarbassov, D. D., Guertin, D. A., Ali, S. M. & Sabatini, D. M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 (2005).
Matsumoto, M., Pocai, A., Rossetti, L., Depinho, R. A. & Accili, D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 6, 208–216 (2007).
Laplante, M. & Sabatini, D. M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012).
Cross, D. A., Alessi, D. R., Cohen, P., Andjelkovich, M. & Hemmings, B. A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378, 785–789 (1995).
Leto, D. & Saltiel, A. R. Regulation of glucose transport by insulin: traffic control of GLUT4. Nat. Rev. Mol. Cell Biol. 13, 383–396 (2012).
Gehart, H., Kumpf, S., Ittner, A. & Ricci, R. MAPK signalling in cellular metabolism: stress or wellness. EMBO Rep. 11, 834–840 (2010).
Cobb, M. H. MAP kinase pathways. Prog. Biophys. Mol. Biol. 71, 479–500 (1999).
Pronk, G. J., McGlade, J., Pelicci, G., Pawson, T. & Bos, J. L. Insulin-induced phosphorylation of the 46- and 52-kDa Shc proteins. J. Biol. Chem. 268, 5748–5753 (1993).
Nichols, R. J. et al. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat. Cell Biol. 20, 1064–1073 (2018).
Ozaki, K. et al. Localization of insulin receptor-related receptor in the rat kidney. Kidney Int. 52, 694–698 (1997).
Vitzthum, H. et al. The AE4 transporter mediates kidney acid-base sensing. Nat. Commun. 14, 3051 (2023).
Elchebly, M. et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 283, 1544–1548 (1999).
Salmeen, A., Andersen, J. N., Myers, M. P., Tonks, N. K. & Barford, D. Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B. Mol. Cell 6, 1401–1412 (2000).
Sevillano, J., Sanchez-Alonso, M. G., Pizarro-Delgado, J. & Ramos-Alvarez, M. D. P. Role of receptor protein tyrosine phosphatases (RPTPs) in insulin signaling and secretion. Int. J. Mol. Sci. 22, 5812 (2021).
Powers, A. C. Type 1 diabetes mellitus: much progress, many opportunities. J. Clin. Invest. 131, e142242 (2021).
Taylor, S. I., Yazdi, Z. S. & Beitelshees, A. L. Pharmacological treatment of hyperglycemia in type 2 diabetes. J. Clin. Invest. 131, e142243 (2021).
McIntyre, H. D. et al. Gestational diabetes mellitus. Nat. Rev. Dis. Primers 5, 47 (2019).
Semple, R. K., Savage, D. B., Cochran, E. K., Gorden, P. & O’Rahilly, S. Genetic syndromes of severe insulin resistance. Endocr. Rev. 32, 498–514 (2011).
Angelidi, A. M., Filippaios, A. & Mantzoros, C. S. Severe insulin resistance syndromes. J. Clin. Invest. 131, e142245 (2021).
Milman, S., Huffman, D. M. & Barzilai, N. The somatotropic axis in human aging: framework for the current state of knowledge and future research. Cell Metab. 23, 980–989 (2016).
Sélénou, C., Brioude, F., Giabicani, E., Sobrier, M. L. & Netchine, I. IGF2: development, genetic and epigenetic abnormalities. Cells 11, 1886 (2022).
Gallagher, E. J. & LeRoith, D. Minireview: IGF, insulin, and cancer. Endocrinology 152, 2546–2551 (2011).
Denduluri, S. K. et al. Insulin-like growth factor (IGF) signaling in tumorigenesis and the development of cancer drug resistance. Genes Dis. 2, 13–25 (2015).
Gallagher, E. J. & LeRoith, D. Hyperinsulinaemia in cancer. Nat. Rev. Cancer 20, 629–644 (2020).
de la Monte, S. M. & Wands, J. R. Review of insulin and insulin-like growth factor expression, signaling, and malfunction in the central nervous system: relevance to Alzheimer’s disease. J. Alzheimers Dis. 7, 45–61 (2005).
Arnold, S. E. et al. Brain insulin resistance in type 2 diabetes and Alzheimer disease: concepts and conundrums. Nat. Rev. Neurol. 14, 168–181 (2018).
Alberini, C. M. IGF2 in memory, neurodevelopmental disorders, and neurodegenerative diseases. Trends Neurosci. 46, 488–502 (2023).
Guarente, L., Sinclair, D. A. & Kroemer, G. Human trials exploring anti-aging medicines. Cell Metab. 36, 354–376 (2024).
Bornfeldt, K. E. & Tabas, I. Insulin resistance, hyperglycemia, and atherosclerosis. Cell Metab. 14, 575–585 (2011).
von der Thüsen, J. H. et al. IGF-1 has plaque-stabilizing effects in atherosclerosis by altering vascular smooth muscle cell phenotype. Am. J. Pathol. 178, 924–934 (2011).
Laron, Z. & Werner, H. Administration of insulin like growth factor I (IGF-I) lowers serum lipoprotein(a)-impact on atherosclerotic cardiovascular disease. Growth Hormone IGF Res. 71, 101548 (2023).
Little, K. et al. Common pathways in dementia and diabetic retinopathy: understanding the mechanisms of diabetes-related cognitive decline. Trends Endocrinol. Metab. 33, 50–71 (2022).
Tsui, S. et al. Evidence for an association between thyroid-stimulating hormone and insulin-like growth factor 1 receptors: a tale of two antigens implicated in Graves’ disease. J. Immunol. 181, 4397–4405 (2008).
Smith, T. J. & Janssen, J. Insulin-like growth factor-I receptor and thyroid-associated ophthalmopathy. Endocr. Rev. 40, 236–267 (2019).
Hales, C. N. & Ozanne, S. E. For debate: fetal and early postnatal growth restriction lead to diabetes, the metabolic syndrome and renal failure. Diabetologia 46, 1013–1019 (2003).
Saenger, P., Czernichow, P., Hughes, I. & Reiter, E. O. Small for gestational age: short stature and beyond. Endocr. Rev. 28, 219–251 (2007).
Veenendaal, M. V. et al. Transgenerational effects of prenatal exposure to the 1944–45 Dutch famine. BJOG 120, 548–553 (2013).
Painter, R. C. et al. Transgenerational effects of prenatal exposure to the Dutch famine on neonatal adiposity and health in later life. BJOG 115, 1243–1249 (2008).
Vogt, M. C. & Hobert, O. Starvation-induced changes in somatic insulin/IGF-1R signaling drive metabolic programming across generations. Sci. Adv. 9, eade1817 (2023).
Voo, K. et al. Maternal starvation primes progeny response to nutritional stress. PLoS Genet. 17, e1009932 (2021).
Zhu, Z. et al. Global histone H2B degradation regulates insulin/IGF signaling-mediated nutrient stress. EMBO J. 42, e113328 (2023).
Burton, N. O. et al. Insulin-like signalling to the maternal germline controls progeny response to osmotic stress. Nat. Cell Biol. 19, 252–257 (2017).
Kajimura, S., Aida, K. & Duan, C. Insulin-like growth factor-binding protein-1 (IGFBP-1) mediates hypoxia-induced embryonic growth and developmental retardation. Proc. Natl Acad. Sci. USA 102, 1240–1245 (2005).
Gupta, M. B., Biggar, K. K., Li, C., Nathanielsz, P. W. & Jansson, T. Increased colocalization and interaction between decidual protein kinase A and insulin-like growth factor-binding protein-1 in intrauterine growth restriction. J. Histochem. Cytochem. 70, 515–530 (2022).
Acknowledgements
This work was supported by NIH grants (R35GM142937 to E.C.; R01GM136976 to X.-C.B.); National Science Foundation grant (IOS-2402404 to CD); the Irma T. Hirschl award (to E.C.); and the Welch foundation (I-1944 to X.-C.B.).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Molecular Cell Biology thanks Andrzej Brzozowski, Jiri Jiracek and the other, anonymous, reviewer for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Choi, E., Duan, C. & Bai, Xc. Regulation and function of insulin and insulin-like growth factor receptor signalling. Nat Rev Mol Cell Biol (2025). https://doi.org/10.1038/s41580-025-00826-3
Accepted:
Published:
DOI: https://doi.org/10.1038/s41580-025-00826-3