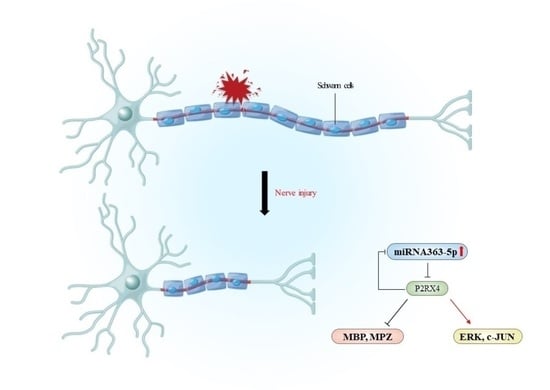
Involvement of the miR-363-5p/P2RX4 Axis in Regulating Schwann Cell Phenotype after Nerve Injury
Abstract
:1. Introduction
2. Results
2.1. Functional Analysis of miRNAs in cAMP-Induced Schwann Cells
2.2. miRNA 363-5p and miRNA 335 Levels are Elevated in Rat Sciatic Nerves during the Postnatal Developmental Period and Decrease after Nerve Injury
2.3. P2RX4 Is a Direct Target of miRNA 363-5p
2.4. P2RX4 Is Elevated in Sciatic Nerve Injury
2.5. P2RX4 Regulates c-JUN and p-ERK Signals after Injury
2.6. P2RX4 and miRNA 363-5p May Be Important Regulators of Myelin Protein Breakdown
2.7. miRNA 363-5p and P2RX4 May Regulate Double-Negative Feedback during Wallerian Degeneration
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Sciatic Nerve Injury
4.3. Peripheral Nerve Development
4.4. Explant Culture
4.5. Semithin Section Analysis
4.6. Intraperitoneal Administration of the P2RX4 Antagonist PSB12062
4.7. Immunofluorescence Staining
4.8. Transfection
4.9. miRNA Array
4.10. Biological Pathway Analyses
4.11. Luciferase Assay
4.12. Real-Time Quantitative PCR
4.13. Western Blotting
4.14. Annotation of miRNA Targets
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, Z.L.; Yu, W.M.; Strickland, S. Peripheral regeneration. Annu. Rev. Neurosci. 2007, 30, 209–233. [Google Scholar] [CrossRef] [PubMed]
- North, R.A. Molecular physiology of P2X receptors. Physiol. Rev. 2002, 82, 1013–1067. [Google Scholar] [CrossRef] [PubMed]
- Khakh, B.S.; North, R.A. P2X receptors as cell-surface ATP sensors in health and disease. Nature 2006, 442, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Ulmann, L.; Hirbec, H.; Rassendren, F. P2X4 receptors mediate PGE2 release by tissue-resident macrophages and initiate inflammatory pain. EMBO J. 2010, 29, 2290–2300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulmann, L.; Hatcher, J.P.; Hughes, J.P.; Chaumont, S.; Green, P.J.; Conquet, F.; Buell, G.N.; Reeve, A.J.; Chessell, I.P.; Rassendren, F. Up-regulation of P2X4 receptors in spinal microglia after peripheral nerve injury mediates BDNF release and neuropathic pain. J. Neurosci. 2008, 28, 11263–11268. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, M.; Shigemoto-Mogami, Y.; Koizumi, S.; Mizokoshi, A.; Kohsaka, S.; Salter, M.W.; Inoue, K. P2X4 receptors induced in spinal microglia gate tactile allodynia after nerve injury. Nature 2003, 424, 778–783. [Google Scholar] [CrossRef]
- Ledderose, C.; Liu, K.; Kondo, Y.; Slubowski, C.J.; Dertnig, T.; Denicolo, S.; Arbab, M.; Hubner, J.; Konrad, K.; Fakhari, M.; et al. Purinergic P2X4 receptors and mitochondrial ATP production regulate T cell migration. J. Clin. Investig. 2018, 128, 3583–3594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, T.; Shen, J.B.; Yang, R.; Redden, J.; Dodge-Kafka, K.; Grady, J.; Jacobson, K.A.; Liang, B.T. Novel protective role of endogenous cardiac myocyte P2X4 receptors in heart failure. Circ. Heart Fail. 2014, 7, 510–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vazquez-Villoldo, N.; Domercq, M.; Martin, A.; Llop, J.; Gomez-Vallejo, V.; Matute, C. P2X4 receptors control the fate and survival of activated microglia. Glia 2014, 62, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.A.; Sun, Q.; Li, Y.C.; Weng, R.X.; Wu, R.; Zhang, H.H.; Xu, G.Y. Overexpression of Purinergic P2X4 Receptors in Hippocampus Rescues Memory Impairment in Rats with Type 2 Diabetes. Neurosci. Bull. 2020, 36, 719–732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macfarlane, L.A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pillai, R.S. MicroRNA function: Multiple mechanisms for a tiny RNA? RNA 2005, 11, 1753–1761. [Google Scholar] [CrossRef] [Green Version]
- Mo, Y.; Liu, B.; Qiu, S.; Wang, X.; Zhong, L.; Han, X.; Mi, F. Down-regulation of microRNA-34c-5p alleviates neuropathic pain via the SIRT1/STAT3 signaling pathway in rat models of chronic constriction injury of sciatic nerve. J. Neurochem. 2020, 154, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.Y.; Miao, Y.; Wang, X.H.; Wang, P.; Cheng, Z.C.; Qian, T.M. Increased levels of miR-3099 induced by peripheral nerve injury promote Schwann cell proliferation and migration. Neural Regen. Res. 2019, 14, 525–531. [Google Scholar] [PubMed]
- Nagata, K.; Hama, I.; Kiryu-Seo, S.; Kiyama, H. microRNA-124 is down regulated in nerve-injured motor neurons and it potentially targets mRNAs for KLF6 and STAT3. Neuroscience 2014, 256, 426–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viader, A.; Chang, L.W.; Fahrner, T.; Nagarajan, R.; Milbrandt, J. MicroRNAs modulate Schwann cell response to nerve injury by reinforcing transcriptional silencing of dedifferentiation-related genes. J. Neurosci. 2011, 31, 17358–17369. [Google Scholar] [CrossRef] [Green Version]
- Bremer, J.; O’Connor, T.; Tiberi, C.; Rehrauer, H.; Weis, J.; Aguzzi, A. Ablation of Dicer from murine Schwann cells increases their proliferation while blocking myelination. PLoS ONE 2010, 5, e12450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, J.A.; Baumann, R.; Norrmén, C.; Somandin, C.; Miehe, M.; Jacob, C.; Lühmann, T.; Hall-Bozic, H.; Mantei, N.; Meijer, D.; et al. Dicer in Schwann Cells is Required for Myelination and Axonal Integrity. J. Neurosci. 2010, 30, 6763–6775. [Google Scholar] [CrossRef] [Green Version]
- Gambarotta, G.; Fregnan, F.; Gnavi, S.; Perroteau, I. Neuregulin 1 role in Schwann cell regulation and potential applications to promote peripheral nerve regeneration. Int. Rev. Neurobiol. 2013, 108, 223–256. [Google Scholar] [PubMed]
- Fricker, F.R.; Bennett, D.L. The role of neuregulin-1 in the response to nerve injury. Future Neurol. 2011, 6, 809–822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoyng, S.A.; de Winter, F.; Tannemaat, M.R.; Blits, B.; Malessy, M.J.; Verhaagen, J. Gene therapy and peripheral nerve repair: A perspective. Front. Mol. Neurosci. 2015, 8, 32. [Google Scholar] [CrossRef] [PubMed]
- Jager, S.B.; Ronchi, G.; Vaegter, C.B.; Geuna, S. The mouse median nerve experimental model in regenerative research. Biomed Res. Int. 2014, 2014, 701682. [Google Scholar] [PubMed]
- Bauder, A.R.; Ferguson, T.A. Reproducible mouse sciatic nerve crush and subsequent assessment of regeneration by whole mount muscle analysis. J. Vis. Exp. 2012, 60, 3606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.; Peng, J.; Han, G.H.; Ding, X.; Wei, S.; Gao, G.; Huang, K.; Chang, F.; Wang, Y. Role of macrophages in peripheral nerve injury and repair. Neural Regen. Res. 2019, 14, 1335–1342. [Google Scholar] [PubMed]
- Jessen, K.R.; Mirsky, R. Negative regulation of myelination: Relevance for development, injury, and demyelinating disease. Glia 2008, 56, 1552–1565. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, D.B.; Bhaskaran, A.; Arthur-Farraj, P.; Noon, L.A.; Woodhoo, A.; Lloyd, A.C.; Feltri, M.L.; Wrabetz, L.; Behrens, A.; Mirsky, R.; et al. c-Jun is a negative regulator of myelination. J. Cell Biol. 2008, 181, 625–637. [Google Scholar] [CrossRef] [Green Version]
- Hernandez-Olmos, V.; Abdelrahman, A.; El-Tayeb, A.; Freudendahl, D.; Weinhausen, S.; Muller, C.E. N-substituted phenoxazine and acridone derivatives: Structure-activity relationships of potent P2X4 receptor antagonists. J. Med. Chem. 2012, 55, 9576–9588. [Google Scholar] [CrossRef]
- Gomez-Sanchez, J.A.; Carty, L.; Iruarrizaga-Lejarreta, M.; Palomo-Irigoyen, M.; Varela-Rey, M.; Griffith, M.; Hantke, J.; Macias-Camara, N.; Azkargorta, M.; Aurrekoetxea, I.; et al. Schwann cell autophagy, myelinophagy, initiates myelin clearance from injured nerves. J. Cell Biol. 2015, 210, 153–168. [Google Scholar] [CrossRef] [Green Version]
- Yi, S.; Wang, Q.H.; Zhao, L.L.; Qin, J.; Wang, Y.X.; Yu, B.; Zhou, S.L. miR-30c promotes Schwann cell remyelination following peripheral nerve injury. Neural Regen. Res. 2017, 12, 1708–1715. [Google Scholar]
- Sohn, E.J.; Park, H.T. MicroRNA Mediated Regulation of Schwann Cell Migration and Proliferation in Peripheral Nerve Injury. Biomed Res. Int. 2018, 2018, 8198365. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Wang, X.; Gu, Y.; Chen, C.; Wang, Y.; Liu, J.; Hu, W.; Yu, B.; Wang, Y.; Ding, F.; et al. Let-7 microRNAs regenerate peripheral nerve regeneration by targeting nerve growth factor. Mol. Ther. 2015, 23, 423–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Liu, Y.; Wu, S.; Zhao, X. Ca2+ Signaling in Oligodendrocyte Development. Cell Mol. Neurobiol. 2019, 39, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Heredia, D.J.; De Angeli, C.; Fedi, C.; Gould, T.W. Calcium Signaling in Schwann cells. Neurosci. Lett. 2020, 729, 134959. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.J.; Yan, Y.; Zhang, L.L.; Agresti, M.A.; Matloub, H.S.; LoGiudice, J.A.; Havlik, R.; Yan, J.G. Increasing Calcium Level Limits Schwann Cell Numbers In Vitro Following Peripheral Nerve Injury. J. Reconstr. Microsurg. 2017, 33, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Itokazu, T.; Hayano, Y.; Takahashi, R.; Yamashita, T. Involvement of Wnt/beta-catenin signaling in the development of neuropathic pain. Neurosci. Res. 2014, 79, 34–40. [Google Scholar] [CrossRef]
- Zhang, Y.K.; Huang, Z.J.; Liu, S.; Liu, Y.P.; Song, A.A.; Song, X.J. WNT signaling underlies the pathogenesis of neuropathic pain in rodents. J. Clin. Investig. 2013, 123, 2268–2286. [Google Scholar] [CrossRef] [PubMed]
- Nickols, J.C.; Valentine, W.; Kanwal, S.; Carter, B.D. Activation of the transcription factor NF-kappa B in Schwann cells is required for peripheral myelin formation. Nat. Neurosci. 2003, 6, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Yoon, C.; Korade, Z.; Carter, B.D. Protein kinase A-induced phosphorylation of the p65 subunit of nuclear factor-kappaB promotes Schwann cell differentiation into a myelinating phenotype. J. Neurosci. 2008, 28, 3738–3746. [Google Scholar] [CrossRef] [Green Version]
- Boyd, J.G.; Gordon, T. Neurotrophic factors and their receptors in axonal regeneration and functional recovery after peripheral nerve injury. Mol. Neurobiol. 2003, 27, 277–323. [Google Scholar] [CrossRef]
- Gordon, T. Neurotrophic factor expression in denervated motor and sensory Schwann cells: Relevance to specificity of peripheral nerve regeneration. Exp. Neurol. 2014, 254, 99–108. [Google Scholar] [CrossRef]
- Fu, S.Y.; Gordon, T. The cellular and molecular basis of peripheral nerve regeneration. Mol. Neurobiol. 1997, 14, 67–116. [Google Scholar] [CrossRef] [PubMed]
- Lalisse, S.; Hua, J.; Lenoir, M.; Linck, N.; Rassendren, F.; Ulmann, L. Sensory neuronal P2RX4 receptors controls BDNF signaling in inflammatory pain. Sci. Rep. 2018, 8, 964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheu, J.Y.; Kulhanek, D.J.; Eckenstein, F.P. Differential patterns of ERK and STAT3 phosphorylation after sciatic nerve transection in the rat. Exp. Neurol. 2000, 166, 392–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agthong, S.; Kaewsema, A.; Tanomsridejchai, N.; Chentanez, V. Activation of MAPK ERK in peripheral nerve after injury. BMC Neurosci. 2006, 7, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monje, P.V.; Bartlett Bunge, M.; Wood, P.M. Cyclic AMP synergistically enhances neuregulin-dependent ERK and Akt activation and cell cycle progression in Schwann cells. Glia 2006, 53, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Arthur-Farraj, P.J.; Latouche, M.; Wilton, D.K.; Quintes, S.; Chabrol, E.; Banerjee, A.; Woodhoo, A.; Jenkins, B.; Rahman, M.; Turmaine, M.; et al. c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron 2012, 75, 633–647. [Google Scholar] [CrossRef] [Green Version]
- Peltonen, S.; Alanne, M.; Peltonen, J. Barriers of the peripheral nerve. Tissue Barriers 2013, 1, e24956. [Google Scholar] [CrossRef] [Green Version]
- Shin, Y.K.; Jang, S.Y.; Park, S.Y.; Park, J.Y.; Kim, J.K.; Kim, J.P.; Suh, D.J.; Lee, H.J.; Park, H.T. Grb2-associated binder-1 is required for neuregulin-1-induced peripheral nerve myelination. J. Neurosci. 2014, 34, 7657–7662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sohn, E.J.; Won, G.; Lee, J.; Lee, S.; Kim, S.H. Upregulation of miRNA3195 and miRNA374b Mediates the Anti-Angiogenic Properties of Melatonin in Hypoxic PC-3 Prostate Cancer Cells. J. Cancer 2015, 6, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Cai, W.; Lee, H.K.; Pellegatta, M.; Shin, Y.K.; Jang, S.Y.; Suh, D.J.; Wrabetz, L.; Feltri, M.L.; Park, H.T. Actin polymerization is essential for myelin sheath fragmentation during Wallerian degeneration. J. Neurosci. 2011, 31, 2009–2015. [Google Scholar] [CrossRef] [PubMed]
- Willis, D.E.; Xu, M.; Donnelly, C.J.; Tep, C.; Kendall, M.; Erenstheyn, M.; English, A.W.; Schanen, N.C.; Kirn-Safran, C.B.; Yoon, S.O.; et al. Axonal Localization of transgene mRNA in mature PNS and CNS neurons. J. Neurosci. 2011, 31, 14481–14487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Shan, Q.; Pan, J.; Yi, S. Actin Cytoskeleton Affects Schwann Cell Migration and Peripheral Nerve Regeneration. Front. Physiol. 2018, 9, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sohn, E.-J.; Nam, Y.-K.; Park, H.-T. Involvement of the miR-363-5p/P2RX4 Axis in Regulating Schwann Cell Phenotype after Nerve Injury. Int. J. Mol. Sci. 2021, 22, 11601. https://doi.org/10.3390/ijms222111601
Sohn E-J, Nam Y-K, Park H-T. Involvement of the miR-363-5p/P2RX4 Axis in Regulating Schwann Cell Phenotype after Nerve Injury. International Journal of Molecular Sciences. 2021; 22(21):11601. https://doi.org/10.3390/ijms222111601
Chicago/Turabian StyleSohn, Eun-Jung, Yun-Kyeong Nam, and Hwan-Tae Park. 2021. "Involvement of the miR-363-5p/P2RX4 Axis in Regulating Schwann Cell Phenotype after Nerve Injury" International Journal of Molecular Sciences 22, no. 21: 11601. https://doi.org/10.3390/ijms222111601