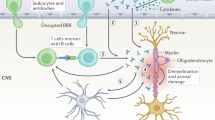
Abstract
Although B cells are implicated in multiple sclerosis (MS) pathophysiology, a predictive or diagnostic autoantibody remains elusive. In this study, the Department of Defense Serum Repository (DoDSR), a cohort of over 10 million individuals, was used to generate whole-proteome autoantibody profiles of hundreds of patients with MS (PwMS) years before and subsequently after MS onset. This analysis defines a unique cluster in approximately 10% of PwMS who share an autoantibody signature against a common motif that has similarity with many human pathogens. These patients exhibit antibody reactivity years before developing MS symptoms and have higher levels of serum neurofilament light (sNfL) compared to other PwMS. Furthermore, this profile is preserved over time, providing molecular evidence for an immunologically active preclinical period years before clinical onset. This autoantibody reactivity was validated in samples from a separate incident MS cohort in both cerebrospinal fluid and serum, where it is highly specific for patients eventually diagnosed with MS. This signature is a starting point for further immunological characterization of this MS patient subset and may be clinically useful as an antigen-specific biomarker for high-risk patients with clinically or radiologically isolated neuroinflammatory syndromes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The PROSITE database used for this study is freely available (https://prosite.expasy.org/). All data produced in the present study are available online at https://github.com/UCSF-Wilson-Lab/MS_DoD_and_ORIGINS_study_data.
Code availability
All analysis code can be found in the following publicly available repository: https://github.com/UCSF-Wilson-Lab/MS_DoD_and_ORIGINS_study_data. PairSeq package source code can be found in the following publicly available repository: https://github.com/cmbartley/PairSeq.
References
Reynolds, R. et al. The neuropathological basis of clinical progression in multiple sclerosis. Acta Neuropathol. 122, 155–170 (2011).
Freedman, M. S. Multiple Sclerosis and Demyelinating Diseases (Lippincott Williams & Wilkins, 2006).
Sospedra, M. & Martin, R. Immunology of multiple sclerosis. Annu. Rev. Immunol. 23, 683–747 (2005).
Lassmann, H., Brück, W. & Lucchinetti, C. F. The immunopathology of multiple sclerosis: an overview. Brain Pathol. 17, 210–218 (2007).
Li, R., Patterson, K. R. & Bar-Or, A. Reassessing B cell contributions in multiple sclerosis. Nat. Immunol. 19, 696–707 (2018).
Sabatino, J. J., Pröbstel, A.-K. & Zamvil, S. S. B cells in autoimmune and neurodegenerative central nervous system diseases. Nat. Rev. Neurosci. 20, 728–745 (2019).
Walton, C. et al. Rising prevalence of multiple sclerosis worldwide: insights from the Atlas of MS, third edition. Mult. Scler. 26, 1816–1821 (2020).
Wallin, M. T. et al. The prevalence of MS in the United States: a population-based estimate using health claims data. Neurology 92, e1029–e1040 (2019).
Rovira, A. et al. A single, early magnetic resonance imaging study in the diagnosis of multiple sclerosis. Arch. Neurol. 66, 587–592 (2009).
Lebrun-Frénay, C. et al. Risk factors and time to clinical symptoms of multiple sclerosis among patients with radiologically isolated syndrome. JAMA Netw. Open 4, e2128271 (2021).
Högg, T. et al. Mining healthcare data for markers of the multiple sclerosis prodrome. Mult. Scler. Relat. Disord. 25, 232–240 (2018).
Disanto, G. et al. Prodromal symptoms of multiple sclerosis in primary care. Ann. Neurol. 83, 1162–1173 (2018).
Hauser, S. L. & Oksenberg, J. R. The neurobiology of multiple sclerosis: genes, inflammation, and neurodegeneration. Neuron 52, 61–76 (2006).
Bjornevik, K. et al. Serum neurofilament light chain levels in patients with presymptomatic multiple sclerosis. JAMA Neurol. 77, 58–64 (2020).
Bjornevik, K. et al. Longitudinal analysis reveals high prevalence of Epstein–Barr virus associated with multiple sclerosis. Science 375, 296–301 (2022).
Ning, L. & Wang, B. Neurofilament light chain in blood as a diagnostic and predictive biomarker for multiple sclerosis: a systematic review and meta-analysis. PLoS ONE 17, e0274565 (2022).
Arbuckle, M. R. et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N. Engl. J. Med. 349, 1526–1533 (2003).
Primavera, M., Giannini, C. & Chiarelli, F. Prediction and prevention of type 1 diabetes. Front. Endocrinol. (Lausanne) 11, 248 (2020).
Alpizar-Rodriguez, D. & Finckh, A. Is the prevention of rheumatoid arthritis possible? Clin. Rheumatol. 39, 1383–1389 (2020).
Hundt, J. E., Hoffmann, M. H., Amber, K. T. & Ludwig, R. J. Editorial: Autoimmune pre-disease. Front. Immunol. 14, 1159396 (2023).
Höftberger, R., Lassmann, H., Berger, T. & Reindl, M. Pathogenic autoantibodies in multiple sclerosis—from a simple idea to a complex concept. Nat. Rev. Neurol. 18, 681–688 (2022).
Kuerten, S. et al. Autoantibodies against central nervous system antigens in a subset of B cell–dominant multiple sclerosis patients. Proc. Natl Acad. Sci. USA 117, 21512–21518 (2020).
McLaughlin, K. A. & Wucherpfennig, K. W. B cells and autoantibodies in the pathogenesis of multiple sclerosis and related inflammatory demyelinating diseases. Adv. Immunol. 98, 121–149 (2008).
Ramesh, A. et al. A pathogenic and clonally expanded B cell transcriptome in active multiple sclerosis. Proc. Natl Acad. Sci. USA 117, 22932–22943 (2020).
Willis, S. N. et al. Investigating the antigen specificity of multiple sclerosis central nervous system-derived immunoglobulins. Front. Immunol. 6, 600 (2015).
Elliott, C. et al. Functional identification of pathogenic autoantibody responses in patients with multiple sclerosis. Brain 135, 1819–1833 (2012).
Greenfield, A. L. et al. Longitudinally persistent cerebrospinal fluid B-cells can resist treatment in multiple sclerosis. JCI Insight 4, e126599 (2019).
Petzold, A. Intrathecal oligoclonal IgG synthesis in multiple sclerosis. J. Neuroimmunol. 262, 1–10 (2013).
Wijnands, J. M. A. et al. Prodrome in relapsing-remitting and primary progressive multiple sclerosis. Eur. J. Neurol. 26, 1032–1036 (2019).
Tremlett, H. & Marrie, R. A. The multiple sclerosis prodrome: emerging evidence, challenges, and opportunities. Mult. Scler. 27, 6–12 (2021).
Pavlin, J. A. & Welch, R. A. Ethics, human use, and the Department of Defense Serum Repository. Mil. Med. 180, 49–56 (2015).
Larman, H. B. et al. Autoantigen discovery with a synthetic human peptidome. Nat. Biotechnol. 29, 535–541 (2011).
Mandel-Brehm, C. et al. Kelch-like protein 11 antibodies in seminoma-associated paraneoplastic encephalitis. N. Engl. J. Med. 381, 47–54 (2019).
Vazquez, S. E. et al. Identification of novel, clinically correlated autoantigens in the monogenic autoimmune syndrome APS1 by proteome-wide PhIP-Seq. eLife 9, e55053 (2020).
Mandel-Brehm, C. et al. Autoantibodies to perilipin-1 define a subset of acquired generalized lipodystrophy. Diabetes 72, 59–70 (2022). db211172.
Mandel-Brehm, C. et al. ZSCAN1 autoantibodies are associated with pediatric paraneoplastic ROHHAD. Ann. Neurol. 92, 279–291 (2022).
Larman, H. B. et al. PhIP-Seq characterization of autoantibodies from patients with multiple sclerosis, type 1 diabetes and rheumatoid arthritis. J. Autoimmun. 43, 1–9 (2013).
Rasquinha, M. T. et al. PhIP-Seq reveals autoantibodies for ubiquitously expressed antigens in viral myocarditis. Biology 11, 1055 (2022).
Rasquinha, M. T., Lasrado, N., Larman, B. H. & Reddy, J. PhIP-Seq analysis reveals autoantibodies for novel antigens in the mouse model of Coxsackievirus B3 infection. J. Immunol. 206, 21.19 (2021).
Lanz, T. V. et al. Clonally expanded B cells in multiple sclerosis bind EBV EBNA1 and GlialCAM. Nature 603, 321–327 (2022).
Ayoglu, B. et al. Anoctamin 2 identified as an autoimmune target in multiple sclerosis. Proc. Natl Acad. Sci. USA 113, 2188–2193 (2016).
Tatsuno, T. & Ishigaki, Y. C-terminal short arginine/serine repeat sequence-dependent regulation of Y14 (RBM8A) localization. Sci. Rep. 8, 612 (2018).
Philipps, D., Celotto, A. M., Wang, Q.-Q., Tarng, R. S. & Graveley, B. R. Arginine/serine repeats are sufficient to constitute a splicing activation domain. Nucleic Acids Res. 31, 6502–6508 (2003).
Richardson, D. N. et al. Comparative analysis of serine/arginine-rich proteins across 27 eukaryotes: insights into sub-family classification and extent of alternative splicing. PLoS ONE 6, e24542 (2011).
Sigrist, C. J. A. et al. New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344–D347 (2013).
Wijnands, J. M. A. et al. Health-care use before a first demyelinating event suggestive of a multiple sclerosis prodrome: a matched cohort study. Lancet Neurol. 16, 445–451 (2017).
Cortese, M. et al. Preclinical disease activity in multiple sclerosis: a prospective study of cognitive performance prior to first symptom. Ann. Neurol. 80, 616–624 (2016).
Khalil, M. et al. Neurofilaments as biomarkers in neurological disorders. Nat. Rev. Neurol. 14, 577–589 (2018).
Disanto, G. et al. Serum neurofilament light: a biomarker of neuronal damage in multiple sclerosis. Ann. Neurol. 81, 857–870 (2017).
Bose, G. et al. Early neurofilament light and glial fibrillary acidic protein levels improve predictive models of multiple sclerosis outcomes. Mult. Scler. Relat. Disord. 74, 104695 (2023).
Jons, D. et al. Axonal injury in asymptomatic individuals preceding onset of multiple sclerosis. Ann. Clin. Transl. Neurol. 9, 882–887 (2022).
Benkert, P. et al. Serum neurofilament light chain for individual prognostication of disease activity in people with multiple sclerosis: a retrospective modelling and validation study. Lancet Neurol. 21, 246–257 (2022).
Häusser-Kinzel, S. & Weber, M. S. The role of B cells and antibodies in multiple sclerosis, neuromyelitis optica, and related disorders. Front. Immunol. 10, 201 (2019).
Sechi, E. et al. Myelin oligodendrocyte glycoprotein antibody-associated disease (MOGAD): a review of clinical and MRI features, diagnosis, and management. Front. Neurol. 13, 885218 (2022).
Wingerchuk, D. M. & Lucchinetti, C. F. Neuromyelitis optica spectrum disorder. N. Engl. J. Med. 387, 631–639 (2022).
Cepok, S. et al. Identification of Epstein–Barr virus proteins as putative targets of the immune response in multiple sclerosis. J. Clin. Invest. 115, 1352–1360 (2005).
Riedhammer, C. & Weissert, R. Antigen presentation, autoantigens, and immune regulation in multiple sclerosis and other autoimmune diseases. Front. Immunol. 6, 322 (2015).
Comabella, M. et al. Increased cytomegalovirus immune responses at disease onset are protective in the long-term prognosis of patients with multiple sclerosis. J. Neurol. Neurosurg. Psychiatry 94, 173–180 (2022).
Bar-Or, A. et al. Epstein–Barr virus in multiple sclerosis: theory and emerging immunotherapies. Trends Mol. Med. 26, 296–310 (2020).
Soldan, S. S. & Lieberman, P. M. Epstein–Barr virus and multiple sclerosis. Nat. Rev. Microbiol. 21, 51–64 (2023).
Wallin, M. T. et al. The Gulf War era multiple sclerosis cohort: age and incidence rates by race, sex and service. Brain 135, 1778–1785 (2012).
Wallin, M. T., Culpepper, W. J., Maloni, H. & Kurtzke, J. F. The Gulf War era multiple sclerosis cohort: 3. Early clinical features. Acta Neurol. Scand. 137, 76–84 (2018).
Thompson, A. J. et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 17, 162–173 (2018).
Zamecnik, C. R. et al. ReScan, a multiplex diagnostic pipeline, pans human sera for SARS-CoV-2 antigens. Cell Rep. Med. 1, 100123 (2020).
Vazquez, S. E. et al. Autoantibody discovery across monogenic, acquired, and COVID-19-associated autoimmunity with scalable PhIP-seq. eLife 11, e78550 (2022).
Acknowledgements
This work was supported by the Valhalla Foundation (S.L.H., M.R.W., J.R.O. and B.A.C.); the Weill Neurohub (C.R.Z.); the Westridge Foundation (M.R.W., H.-C.v.B. and A.J.G.); National Institute of Neurological Disorders and Stroke R35NS122073 (S.L.H., M.R.W. and R.D.); National Institute of Allergy and Infectious Diseases R01AI170863-01A1 (S.S.Z.); National Multiple Sclerosis Society RFA-2104-37504 (M.R.W., M.T.W., J.H., B.A.C., C.R.Z., R.D. and R.D.B.); National Multiple Sclerosis Society RFA-2104-3747 (J.R.O.); the Department of Defense (A.A.); the German Society of Multiple Sclerosis (A.A.); the Water Cove Charitable Foundation (M.R.W., M.T.W., J.H., B.A.C. and C.R.Z.); Tim and Laura O’Shaughnessy (M.R.W.); the Littera Family (M.R.W.); the School of Medicine Dean’s Yearlong Fellowship, supported by residual funds from the Howard Hughes Medical Institute Medical Fellows at UCSF (G.M.S.); the Chan Zuckerberg Biohub San Francisco (J.L.D. and S.A.M.); the John A. Watson Scholar Program at UCSF (C.M.B.); the Hanna H. Gray Fellowship, Howard Hughes Medical Institute (C.M.B.); National Institutes of Health R01AI158861 (J.A.H. and K.J.W.); and the University of California President’s Postdoctoral Fellowship Program (C.M.B.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. The authors would like to acknowledge the Chan Zuckerberg Biohub San Francisco and its sequencing team for assistance. The following members of the Walter Reed National Military Medical Center Neurology Department were collaborators on this project: A. Lewis and N. Tagg. Data were obtained from the Defense Medical Surveillance System, Armed Forces Health Surveillance Branch, Defense Health Agency (data from 1987–2007, released 2010). Figures 1 and 5a were created with BioRender. The authors would also like to thank the patients and their families for their contribution to this study.
Author information
Authors and Affiliations
Contributions
C.R.Z., G.M.S., M.R.W., M.T.W. and J.L.D. designed the experiments. C.R.Z. performed all PhIP-Seq and Luminex assays. R.D.B. and C.M.C. performed the Luminex assays, and G.M.S. and S.A.M. performed PhIP-Seq immunoprecipitations and NGS library preparation. S.A.M., C.F. and A.E.W. assisted with DoDSR sample preparation. A.A., N.J. and K.A. performed the sNfL studies, and A.A. analyzed the data. R.D. and C.M.B. built the analysis code and pipeline for PhIP-Seq data and analyzed the PhIP-Seq data, with G.M.S., as well as generated figures. K.C.Z., A.T., C.F., J.A., C.G., E.B.T., H.-C.v.B., R.P.L., R.G. and E.L.E. curated and collected patient samples and clinical data for the UCSF ORIGINS cohort. K.J.W., J.R.O., D.G.A., K.K. and J.A.H. performed the HLA sequencing, genotyping and association. M.T.W. generated the DoDSR cohort and epidemiologic analysis, and A.J.G. contributed valuable sNfL analysis interpretation. S.L.H., B.A.C.C. and J.R.O., together with the UCSF MS-EPIC Team and J.M.G., C.-Y.G. and R.C., led the recruitment and phenotyping of the UCSF ORIGINS cohort. S.L.H., B.A.C.C. and J.L.D. provided valuable experimental guidance and manuscript editing. All authors contributed to study design. C.R.Z., G.M.S. and M.R.W. wrote the manuscript. All authors provided editorial comments.
Corresponding author
Ethics declarations
Competing interests
M.R.W. receives unrelated research grant funding from Roche/Genentech and Novartis and has received speaking honoraria from Genentech, Takeda, WebMD and Novartis. M.R.W. and J.L.D. receive licensing fees from CDI Labs. C.M.B. serves as a paid consultant for the Neuroimmune Foundation. J.J.S. has unrelated research grant funding from Roche/Genentech and Novartis and advisory board honoraria from IgM Biosciences. C.-Y.G. has received financial compensation from serving on advisory boards for Genentech and Horizon. R.G.H. has unrelated research funding from Roche/Genentech and Atara Bio; consulting fees from Roche/Genentech, Novartis and QIA Consulting; and discussion leader fees from Sanofi/Genzyme. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Medicine thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Jerome Staal, in collaboration with the Nature Medicine team
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1
Heatmap of logRPK values from whole proteome. PhIP-Seq data on MS patient sera from DoDSR cohort. Top peptides shown with peptide enrichment. blowout to demonstrate similarity between pre and post onset sample enrichments over time.
Extended Data Fig. 2
We investigated the similarity between different timepoints from the from the DoDSR cohort (n = 500). (Top Left) Unfiltered normalized read counts (rpK) were log transformed, and a correlation matrix was computed using the Euclidean distance measure. The similarity between any two samples was defined by their n-dimensional Euclidean distance, where each peptide in the library was treated as a dimension. The rows and columns of this matrix are ordered such that the early timepoints are followed by the late timepoints. The central diagonal stripe in the heatmap represents a distance of zero where the distance was computed between the same sample. However, the diagonal stripes represent distances calculated between different timepoints of the same sample. (Top Right) Box plots of the Euclidean distance between timepoints, with a trend towards increasing distance at longer intervals. However, patterns of self-reactivity remain stable for more than 15 years. Notches approximate 95% confidence intervals. (Bottom) A boxplot demonstrates the significant difference (two-sided t-test) in Euclidean distance between two timepoints of the same sample versus the distance between two unrelated samples. Notches approximate 95% confidence intervals.
Extended Data Fig. 3
A subset of the library capturing relevant motifs from previously identified putative targets of molecular mimicry such as GlialCAM and anoctamin-2. (Top) MS patient samples are ordered so as to preserve the IC clusters. Although individual samples can be seen to enrich peptides of interest, they did not meet the predefined cutoffs for this analysis. (Bottom) HC samples demonstrating background levels of enrichment.
Extended Data Fig. 4
Time point specific NfL data from DoD cohort. (1st time point: HC n = 236, MSno-IC n = 204, MSIC n = 13. 2nd time point: HC n = 234, MSno-IC n = 191, MSIC n = 21. Data represented as geometric mean and standard factors for each box and whisker.).
Extended Data Fig. 5
Additional OND CSF controls (n = 20), lacking enrichment of the IC motif.
Extended Data Fig. 6
Correlation of sum of MFI from all six peptides of interest in Table 1 from Luminex assay with their sum of respective normalized read counts from PhIP-Seq data in both CSF and serum samples from ORIGINS cohort. Shaded area represents 95% confidence intervals.
Extended Data Fig. 7
Luminex assay for antibodies against listed pathogen peptides in ORIGINS cohort a) MS patient CSF (n = 104), b) MS patient serum (n = 104), c) OND patient CSF(n = 42) and d) OND patient serum (n = 22). e) Same patient populations respresnted as sum of MFI combining signal across pathogen peptides with highlighted patients having normalized sumMFI values > 3 in CSF.
Supplementary information
Supplementary Tables 1 and 2
Supplementary Table 1: HLA data for the ORIGINS cohort; Supplementary Table 2: HLA allele frequency assessment.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zamecnik, C.R., Sowa, G.M., Abdelhak, A. et al. An autoantibody signature predictive for multiple sclerosis. Nat Med 30, 1300–1308 (2024). https://doi.org/10.1038/s41591-024-02938-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-024-02938-3