Results support that the PF sampling method is capable of detecting one single PRRS virus-positive pig among a high number of negative pigs.
December 31, 2018

By Will A. López, Phil Gauger, Karen Harmon, Laura Bradner, Gustavo Silva and Daniel C. L. Linhares, Iowa State University
Processing fluids monitoring is with no doubt a robust tool for porcine reproductive and respiratory syndrome virus monitoring and surveillance in piglets, for sow farms undergoing PRRS virus control/elimination programs, and it appears to be highly sensitive in low prevalence scenarios. The dramatic increase in laboratory submissions of PF samples, is evidence of the high perceived value that the swine industry sees in this population-based sampling technique. Pooling PF for polymerase chain reaction based testing has been documented showing that when litters are pooled by farrowing room (all litters processed within a farrowing room), or by whole day of collection (all litters processed in a day) the performance of the PF method is superior than when PF is tested individually per each litter.
Using aggregated samples such as PF brings the question about the detrimental effect of pooling a sample type that is already in essence, a pool, on the probability of PRRS virus detection by quantitative PCR, especially when prevalence is low. Thus, our objective with this study was to assess the effect of pooling PF on the probability of PRRS virus RNA detection by qPCR under a low prevalence scenario.
Study design
Results of a previous study were analyzed to understand the patterns of PF’s cycle threshold values and their relationship to the number of PCR-positive pigs within PCR-positive litters. It revealed that Ct values of PF from litters having a single viremic piglet had a mean Ct value of 30.52. We called those litters: ‘low prevalence litters’. Thus, a field PF sample previously tested with a Ct value of 28 was selected to represent a litter of 11 pigs having a single viremic piglet (Sample PF1). The sample was used to represent a conservative scenario and to account for an expected increase in Ct value after a freeze-thaw cycle.
Using PF from PRRS-naïve herds, we performed six replications of eight two-fold serial dilutions on the PF1 sample to model diluting PRRS-positive PF (PF1) with PRRS virus-negative PF and gradually increasing the volume of negative PF until losing detectability of PRRS virus RNA by qPCR. For each two-fold dilution, it was considered that the number of PRRS-negative pigs represented increased by a factor of two. For instance, the first dilution would represent a PF from 22 pigs with one viremic piglet. After eight dilutions, the PF would represent 2,816 pigs with one viremic pig.
All samples were tested individually for PRRS virus RNA by qPCR using the commercial RealPCR* IDEXX PRRS kit to determine the probability of PRRS virus RNA detection as a function of the number of PRRS virus-negative pigs contributing to the processing fluid sample in each dilution. The statistical analysis was done using regression models on SAS 9.4.
Results and conclusions
The sample PF1 (initially selected PRRS virus-positive PF with Ct of 28), yielded PRRS virus qPCR results with a mean Ct value of 29.05 from the six replications. The results of qPCR on the serially diluted PF1 sample showed an average increment of 1.37 points in Ct for each two-fold dilution, as expected. The raw probability of detecting PRRS virus by qPCR decreased to 33% at the 6th dilution (352 piglets) then to 17% at the 7th dilution (1408 piglets) and 0% at the 8th dilution (2816 piglets).
Results support that the PF sampling method is capable of detecting one single PRRS virus-positive pig among a high number of negative pigs. The sample PF1 models ‘worst case scenario’ of having just ‘low prevalence litter’ in the PF sample. The regression coefficients for the serially diluted PF1 data indicate that one viremic pig with a Ct value of 29 could be detected in a pooled PF sample with up to 780 PRRS virus-negative pigs, but, what is the probability of that happening?
Figure 1 shows the predicted probabilities for PRRS virus detection by qPCR in case of having one viremic piglet in the pooled sample and points to three different examples of PF pool sizes with its corresponding chances to detect the virus. When there was a single ‘low prevalence litter’ in a room, there was 95%, 80% and 60% probability to detect PRRS virus by qPCR when pooling 25, 48 and 71 litters respectively.
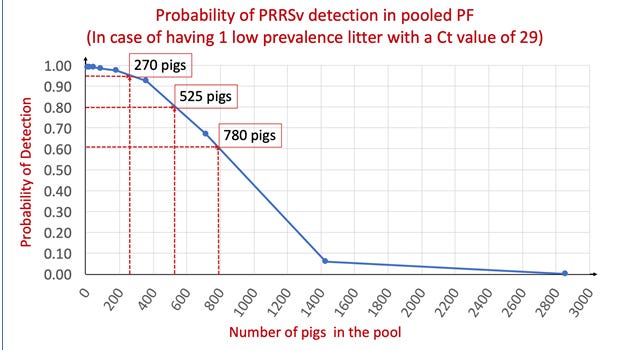
There is a marked dilution effect due to the increasing pooling of PRRS virus-negative PF with the original ‘low prevalence litter’ sample. However, pooling PF (e.g. whole-room as opposed to PF from few litters) would increase the coverage (number of pigs included in the PF sample) which in turn may increase the probability of PRRS virus detection by qPCR. Thus, the ability to pool large numbers of pigs in a PF sample allows to test more pigs more frequently and therefore improve the performance of PRRS virus monitoring and surveillance programs.
References
1. Yeske P. Application of new tools to declare a herd as PRRS negative. 2018.
2. Trevisan G. Swine disease reporting system: Real-time, aggregated results from major U.S. veterinary diagnostic laboratories. 2018; Ames, IA. p 43-44.
3. López W. Processing fluids sensitivity and specificity for RT-PCR assays to detect PRRS virus under field conditions. 2018.
You May Also Like



