(I have decided to try, from time to time, to write about new publications in research on male breast cancer, and give a brief lay explanation of the findings and perhaps a comment. Here goes.)
Combined microRNA and ER expression: a new classifier for familial and sporadic breast cancer patients
by Danza et al. Journal of Translational Medicine 2014, 12:319
Background
Historically cancers were classified simply according to their appearance down the microscope, by the pathologist. Then came some markers, such as the hormone receptors ER and PR, and the growth factor receptor Her2 for breast cancer. These markers have both prognostic value and allow the selection of appropriate therapies. At present development of new drugs goes hand in hand with developing ways to further subdivide tumors into classes based on molecular patterns: cancers that look similar down the microscope, might be divided meaningfully on the basis of molecular markers.
There are many molecular markers you can choose. Dominant for a long time were changes in the genome such as amplifications, deletions and rearrangements of chromosomes. Now with fast affordable sequencing we have growing catalogs of point mutations. More recently the discovery of so called microRNAs, which are regulators of the genome, we have a new tool for classifying. Danza et al are attempting to use microRNAs to make meaningful classifications of familial breast cancers and sporadic breast cancers, in an attempt to learn about biological commonalities, which could eventually be exploited for therapy.
Findings
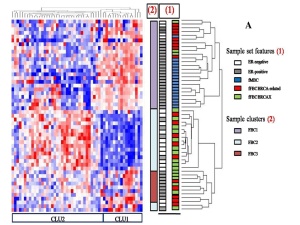
They find three broad clusters based on the expression of microRNAs, and they speculate on their biological significance. Their concluding remarks read in part:
In conclusion, miRNAs expression pattern in familial and sporadic BCs, related to immunophenotype, could better clarify similarities and differences between these two groups.
In other words, the differences that matter (for the purposes of treatment selection) aren’t whether its an inherited or sporadic form of breast cancer, but what its molecular make up is. Makes sense.
Comment
What I found interesting for analysis of the male disease is that all the male samples clustered in one group, which was overwhelmingly ER+ and had mostly BRCA-related female familial breast cancers. This matches what is know about the sporadic cancers – men mostly have ER+/PR+ cancers. It suggests that most (maybe almost all?) male cancers are of a specific subtype, which is important information for assigning the right treatment as (hopefully) new therapies are developed.
I want to express my thanks to the authors for including male breast cancers in their sample set – and not just a few. The work incorporated 39 female familial breast cancer cases (19 BRCA-related and 20 BRCAX) and 12 male (BRCAX) – thats about 30% male samples!